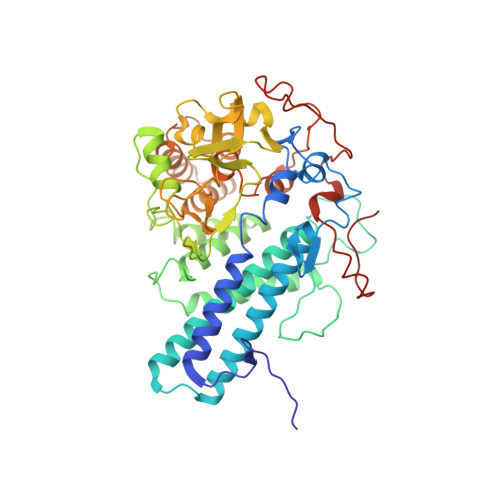
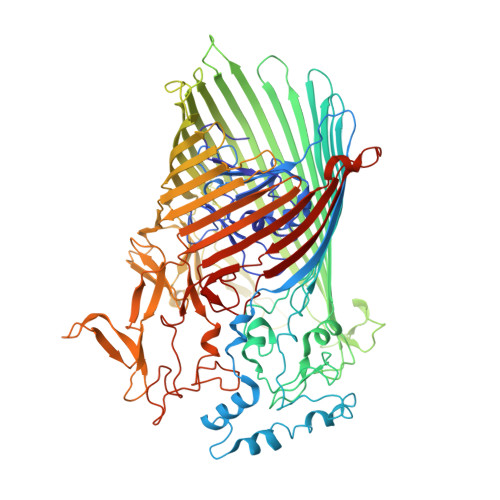
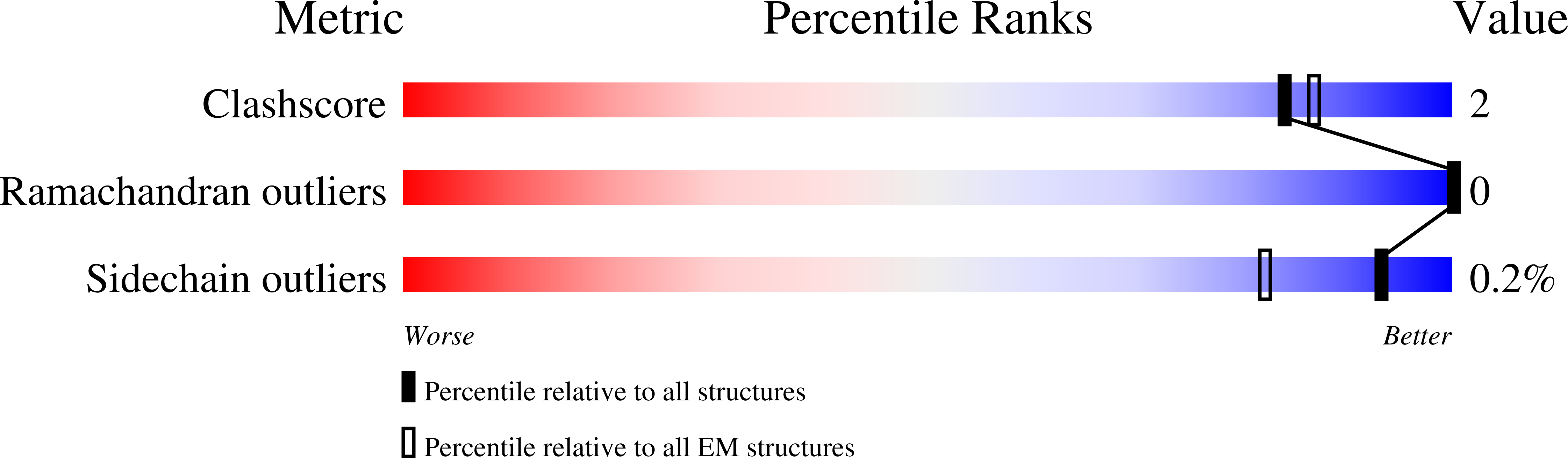Structural and functional insights into oligopeptide acquisition by the RagAB transporter from Porphyromonas gingivalis.
Madej, M., White, J.B.R., Nowakowska, Z., Rawson, S., Scavenius, C., Enghild, J.J., Bereta, G.P., Pothula, K., Kleinekathoefer, U., Basle, A., Ranson, N.A., Potempa, J., van den Berg, B.(2020) Nat Microbiol 5: 1016-1025
- PubMed: 32393857
- DOI: https://doi.org/10.1038/s41564-020-0716-y
- Primary Citation of Related Structures:
6SLI, 6SLJ, 6SLN, 6SM3, 6SML, 6SMQ - PubMed Abstract:
Porphyromonas gingivalis, an asaccharolytic member of the Bacteroidetes, is a keystone pathogen in human periodontitis that may also contribute to the development of other chronic inflammatory diseases. P. gingivalis utilizes protease-generated peptides derived from extracellular proteins for growth, but how these peptides enter the cell is not clear. Here, we identify RagAB as the outer-membrane importer for these peptides. X-ray crystal structures show that the transporter forms a dimeric RagA 2 B 2 complex, with the RagB substrate-binding surface-anchored lipoprotein forming a closed lid on the RagA TonB-dependent transporter. Cryo-electron microscopy structures reveal the opening of the RagB lid and thus provide direct evidence for a 'pedal bin' mechanism of nutrient uptake. Together with mutagenesis, peptide-binding studies and RagAB peptidomics, our work identifies RagAB as a dynamic, selective outer-membrane oligopeptide-acquisition machine that is essential for the efficient utilization of proteinaceous nutrients by P. gingivalis.
Organizational Affiliation:
Department of Microbiology, Faculty of Biochemistry, Biophysics and Biotechnology, Jagiellonian University, Krakow, Poland.