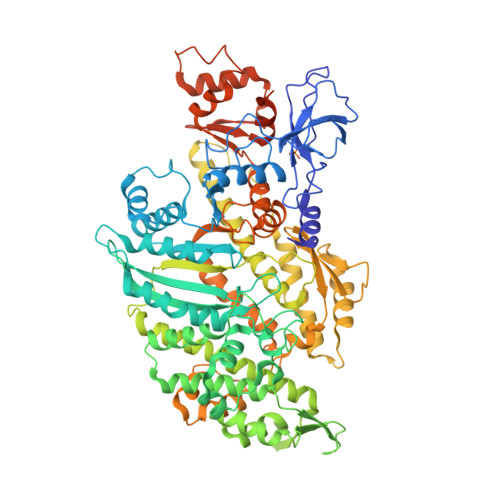
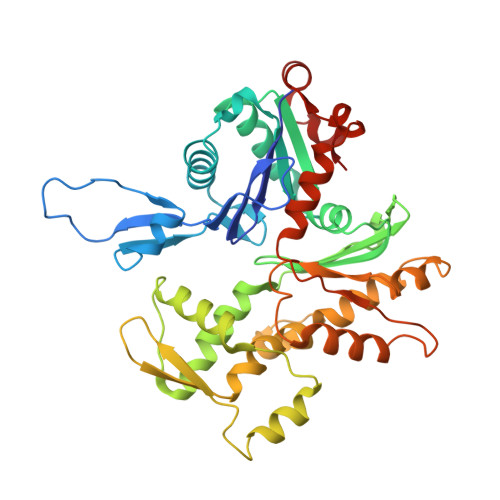
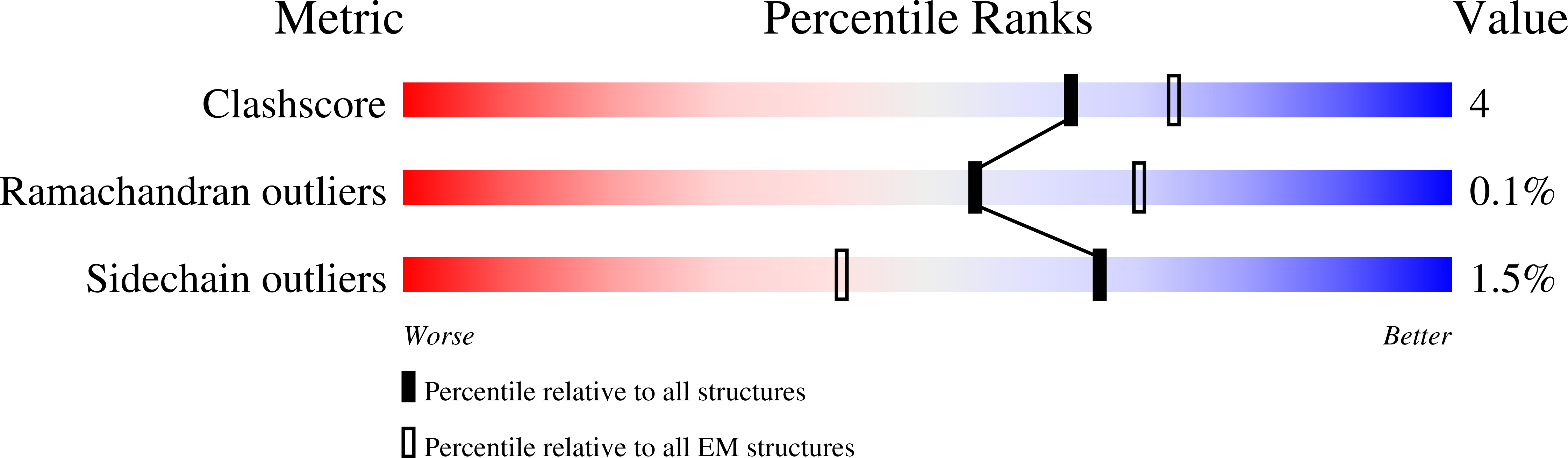High-resolution structures of malaria parasite actomyosin and actin filaments.
Vahokoski, J., Calder, L.J., Lopez, A.J., Molloy, J.E., Kursula, I., Rosenthal, P.B.(2022) PLoS Pathog 18: e1010408-e1010408
- PubMed: 35377914
- DOI: https://doi.org/10.1371/journal.ppat.1010408
- Primary Citation of Related Structures:
6TU4, 6TU7 - PubMed Abstract:
Malaria is responsible for half a million deaths annually and poses a huge economic burden on the developing world. The mosquito-borne parasites (Plasmodium spp.) that cause the disease depend upon an unconventional actomyosin motor for both gliding motility and host cell invasion. The motor system, often referred to as the glideosome complex, remains to be understood in molecular terms and is an attractive target for new drugs that might block the infection pathway. Here, we present the high-resolution structure of the actomyosin motor complex from Plasmodium falciparum. The complex includes the malaria parasite actin filament (PfAct1) complexed with the class XIV myosin motor (PfMyoA) and its two associated light-chains. The high-resolution core structure reveals the PfAct1:PfMyoA interface in atomic detail, while at lower-resolution, we visualize the PfMyoA light-chain binding region, including the essential light chain (PfELC) and the myosin tail interacting protein (PfMTIP). Finally, we report a bare PfAct1 filament structure at improved resolution.
Organizational Affiliation:
Department of Biomedicine, University of Bergen, Bergen, Norway.