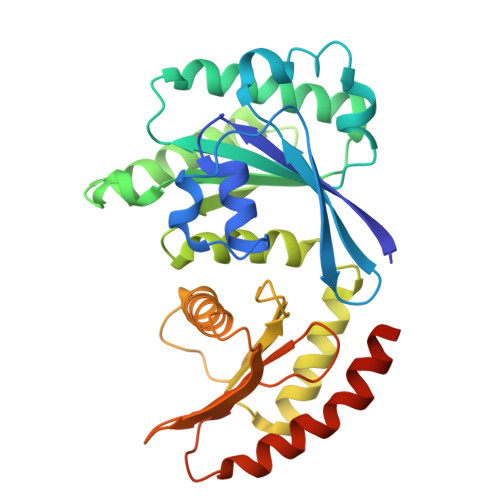
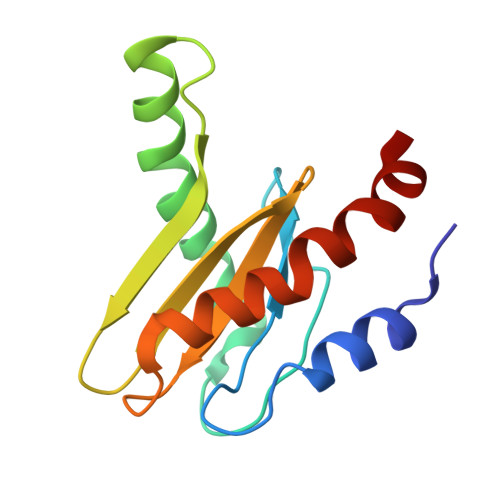
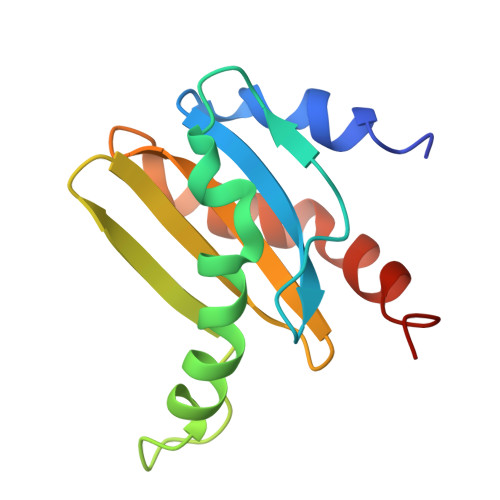
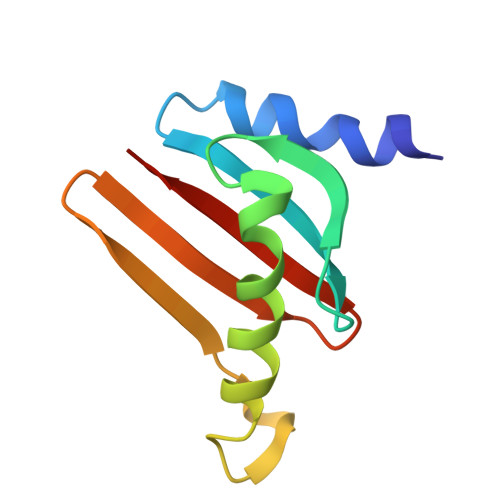
Structural basis for mTORC1 activation on the lysosomal membrane.
Cui, Z., Esposito, A., Napolitano, G., Ballabio, A., Hurley, J.H.(2025) Nature
- PubMed: 40963021
- DOI: https://doi.org/10.1038/s41586-025-09545-3
- Primary Citation of Related Structures:
9ED4, 9ED6, 9ED7, 9ED8 - PubMed Abstract:
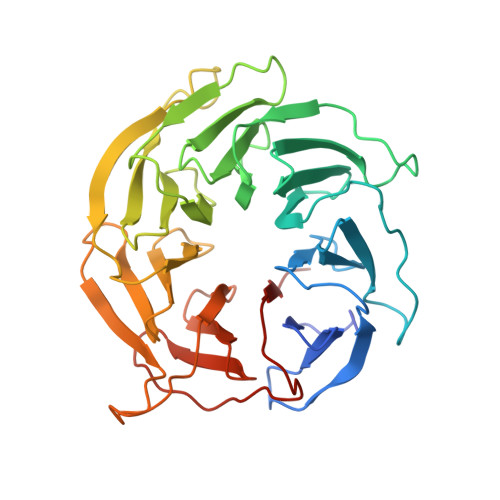
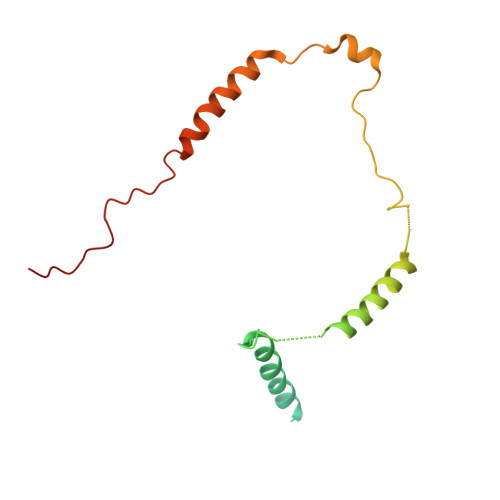
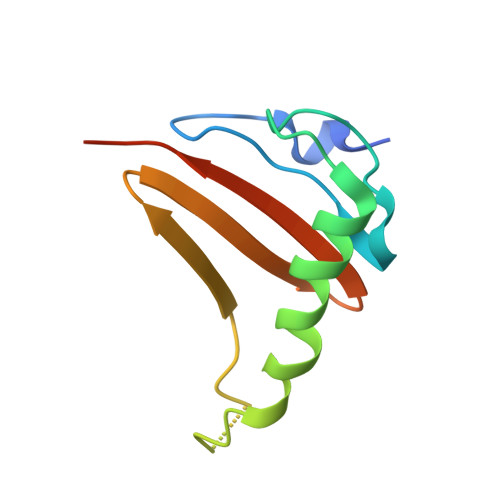
The mechanistic target of rapamycin complex 1 (mTORC1) integrates growth factor (GF) and nutrient signals to stimulate anabolic processes connected to cell growth and inhibit catabolic processes such as autophagy 1,2 . GF signalling through the tuberous sclerosis complex regulates the lysosomally localized small GTPase RAS homologue enriched in brain (RHEB) 3 . Direct binding of RHEB-GTP to the mTOR kinase subunit of mTORC1 allosterically activates the kinase by inducing a large-scale conformational change 4 . Here we reconstituted mTORC1 activation on membranes by RHEB, RAGs and Ragulator. Cryo-electron microscopy showed that RAPTOR and mTOR interact directly with the membrane. Full engagement of the membrane anchors is required for optimal alignment of the catalytic residues in the mTOR kinase active site. Converging signals from GFs and nutrients drive mTORC1 recruitment to and activation on lysosomal membrane in a four-step process, consisting of (1) RAG-Ragulator-driven recruitment to within ~100 Å of the lysosomal membrane; (2) RHEB-driven recruitment to within ~40 Å; (3) RAPTOR-membrane engagement and intermediate enzyme activation; and (4) mTOR-membrane engagement and full enzyme activation. RHEB and membrane engagement combined leads to full catalytic activation and structurally explains GF and nutrient signal integration at the lysosome.
- Department of Molecular and Cell Biology, University of California, Berkeley, Berkeley, CA, USA.
Organizational Affiliation: