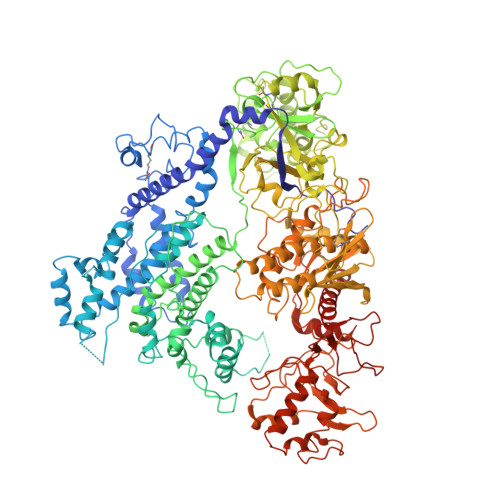
An anti-CRISPR protein disables type V Cas12a by acetylation.
Dong, L., Guan, X., Li, N., Zhang, F., Zhu, Y., Ren, K., Yu, L., Zhou, F., Han, Z., Gao, N., Huang, Z.(2019) Nat Struct Mol Biol 26: 308-314
- PubMed: 30936526
- DOI: https://doi.org/10.1038/s41594-019-0206-1
- Primary Citation of Related Structures:
6IUF, 6IV6 - PubMed Abstract:
Phages use anti-CRISPR proteins to deactivate the CRISPR-Cas system. The mechanisms for the inhibition of type I and type II systems by anti-CRISPRs have been elucidated. However, it has remained unknown how the type V CRISPR-Cas12a (Cpf1) system is inhibited by anti-CRISPRs. Here we identify the anti-CRISPR protein AcrVA5 and report the mechanisms by which it inhibits CRISPR-Cas12a. Our structural and biochemical data show that AcrVA5 functions as an acetyltransferase to modify Moraxella bovoculi (Mb) Cas12a at Lys635, a residue that is required for recognition of the protospacer-adjacent motif. The AcrVA5-mediated modification of MbCas12a results in complete loss of double-stranded DNA (dsDNA)-cleavage activity. In contrast, the Lys635Arg mutation renders MbCas12a completely insensitive to inhibition by AcrVA5. A cryo-EM structure of the AcrVA5-acetylated MbCas12a reveals that Lys635 acetylation provides sufficient steric hindrance to prevent dsDNA substrates from binding to the Cas protein. Our study reveals an unprecedented mechanism of CRISPR-Cas inhibition and suggests an evolutionary arms race between phages and bacteria.
Organizational Affiliation:
HIT Center for Life Sciences, School of Life Science and Technology, Harbin Institute of Technology, Harbin, China.